DNA methylation analysis for the treatment of meningiomas
In 2016, the WHO newly revised the classification system for CNS tumors to support the development of an integrated approach that incorporates both histological examination and molecular genetic analysis in diagnosing patients. While most WHO grade I tumors are benign, it is poorly understood why a very small percentage of these tumors progress. WHO grade II meningiomas have a higher rate of progression than WHO grade I tumors, and they also have a higher rate of recurrence after surgery. The potential for progression or recurrence of either WHO grade I or WHO grade II tumors cannot be accurately predicted by histological examination alone, and, therefore, it is important to identify molecular markers that can be used to predict which tumors are more likely to progress to malignancy or recur after treatment. In addition to predicting meningioma progression and recurrence rates, molecular markers can be used to allow physicians to devise targeted therapy to their specific genetic derangements similar to what is being done with glioma. Recent academic work has made excellent progress in identifying molecular markers that can be used for the genetic profiling of meningiomas. We will provide background and discuss the implications of a new study published by Sahm et al. that proposes a new prognostic classification system for meningiomas based off of DNA methylation patterns (1) (Table 1).
Full table
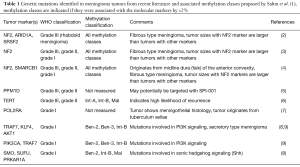
Initial studies have found that NF2 is one of the most commonly found mutated genes in meningiomas (10). More aggressive anaplastic WHO grade III tumors are likely have mutations in NF2 (10), and meningiomas with mutations in NF2 are found to have significantly lower recurrence-free survival rates (3). Sizes of tumors with NF2 mutations were found to be significantly larger than tumors that do not have mutations in NF2. It has been found that 80% of tumors in the calvarium were found to have mutations in NF2, and meningiomas with both mutations in SMARCB1 and NF2 were found to be frequently located in the falx cerebri of patients (3,4). Currently, no targeted therapy exists for NF2 mutations in meningiomas, however, Shah et al. have found that NF2 mutations in ovarian cancer can be successfully targeted by FAK inhibitors (11). In the future, these inhibitors could be further investigated for their use as targeted therapy in meningiomas with mutations in NF2.
In addition to NF2, other mutations have been found in commonly known molecular signaling pathways. Mutations affecting PI3K signaling associated with meningiomas include mutations in AKT1 and PIK3CA. These mutations in PI3K signaling were found to be associated with WHO grade I tumors (7). Studies have also consistently found mutations that affect sonic hedgehog signaling such as mutations in PRKAR1A, SMO and SUFU (7). Meningiomas with mutations in sonic hedgehog signaling were found to be present in WHO grade I tumors that had a low possibility of progressing to malignancy or recurrence after treatment (7). Yuzawa et al. suggest that mutations such as SMO and AKT1 can potentially be targeted by drugs such as hedgehog inhibitors and AKT inhibitors, respectively, if the tumor is in a position that makes it difficult to completely resect (3). PI3K inhibitors could also potentially be used in meningiomas with activation mutations in PI3K (8).
More mutations unrelated to signaling pathways and NF2 have also been discovered. Interestingly, Clark et al. has most recently shown that mutations in POLR2A, a gene in the beta-1 subunit of RNA polymerase II, are indicative of a distinct subset of WHO grade I meningiomas (7). Mutations in RNA polymerase have not been found in any other human pathologies (7). Mutations in TRAF7 and KLF were found to be indicative of secretory meningiomas, which are benign WHO grade I meningiomas characterized by glandular differentiation, perifocal brain edema and pseudopsammoma bodies (9).
Many mutations were found to be indicative of progression to higher grade meningiomas. Mutations in ARID1A, which encodes a subunit of the SWI/SNF complex involved in transcriptional activation of genes, were found to be a predictive marker for the transformation of a meningioma into the rare rhabdoid meningioma. This is a much more malignant form of a WHO grade III meningioma that often carries a poor prognosis (2). Fukami et al. found that overexpression of PPM1D is indicative of progression to either a WHO grade II or grade III meningioma (5). They suggest that PPM1D should be targetable in these tumors, as the small molecule SPI-001 has shown to inhibit PPM1D overexpression in human breast cancer cells (5). Finally, mutations in TERT are commonly found in tumors that progress to WHO grade II or III (6).
In addition to the search for relevant molecular markers, progress has also been made in creating systems that can deliver a faster method for sequencing tumor samples. Yuzawa et al. has produced the first clinical sequencing system for meningiomas that can be completed within 7 days turnaround time for the clinician while using targeted amplicon sequencing (3). This system can identify mutations in NF2, TRAF7, AKT1, KLF4, and SMO, as well as NF2 (3).
These recent studies for identifying mutations in meningiomas have been, so far, unable to sufficiently measure clinical outcomes associated with meningioma mutations to give an accurate indication of a patient’s progression-free survival time. In The Lancet Oncology, Sahm and colleagues present a new classification system that successfully measures DNA methylation in tumor samples to give a more accurate indication of progression-free survival and post-resection recurrence rates of meningiomas than the current WHO grading system (1).
In a retrospective analysis measuring DNA methylation of tumor samples from a large patient population of 490, they discover six methylation classifications that are common in meningiomas. Three classifications named MC ben-1, MC ben-2, and MC ben-3 were designated as methylation classes for benign tumors, and the two methylation classes of MC int-A and MC int-B were designated as methylation classes for intermediate level tumors with a higher rate of progression to malignancy and a higher chance of recurrence after resection. The final methylation class of MC mal (malignant) includes tumors with the highest probability of malignancy and recurrence. In addition to giving clinicians a more accurate prognostication of progression-free survival and rates of recurrence in patients, this system also gives clinicians the ability to distinguish meningiomas on a molecular level from other intracranial tumors that may appear very histologically similar to meningiomas. In their study, they show their methylation system can accurately differentiate meningioma from solitary fibrous tumors, hemangiopericytoma, schwannoma, malignant peripheral nerve sheath tumors, chordoma, chondrosarcoma, hemangioblastoma, fibrous dysplasia, gliosarcoma, leiomyosarcoma, neurofibroma, and fibromatosis.
This method of classification based on DNA methylation patterning is most beneficial for treating WHO grade III and grade II tumors, as these most often require decisions for complex treatment plans and further postoperative monitoring and therapy. A majority of the WHO grade III tumors were found to be in the MC mal methylation class. Additionally, a portion of the grade III tumors were classified as MC int-B (intermediate), and tumors in this methylation classification were shown to have longer progression-free survival times. Patients with meningiomas classified MC int-B had a 50% probability of being progression free at 24 months compared to patients with meningiomas classified MC mal that had a 15.4% probability of being progression free at 24 months. This can give clinical teams, patients and family members a better indication of survival times for patients with grade III tumors, and it will be useful in determining and evaluating optimal new treatment regimens including experimental chemotherapy, fractionated radiotherapy and peptide receptor radionuclide therapy.
This system allows clinicians to better identify which WHO grade II tumors have a higher chance of progression and which tumors have a higher chance of remaining benign. Most of the WHO grade II tumors were found in the methylation classes of MC int-A (intermediate) and MC int-B (intermediate), and one subset was also found in the MC ben-1 (benign) methylation class. Patients with WHO grade II tumors assigned to MC ben-1 had a 75.7% probability of being progression-free at 72 months compared to a 28.3% and 23.1% chance of progression-free at 72 months in both the MC int-A and MC int-B respectively. In WHO grade II meningiomas, fractionated radiotherapy should be used for postoperative treatment in tumors that progress or recur after surgery (10). Methylation classification will assist physicians in making decisions on radiotherapy in WHO grade II tumors, as meningiomas with the MC int-A and MC int-B should be strongly considered for fractionated radiotherapy after the initial surgery.
In the future, methylation classifications can be used to predict targeted therapy treatments for both WHO grade III and WHO grade II meningiomas. Mutations in the sonic hedgehog signaling gene SUFU, a negative regulator of the activation of several target genes, are found in both MC int-B and MC mal. Therefore, GLI inhibitors can be investigated for their use in inhibiting hedgehog signaling in both WHO grade III and WHO grade II meningiomas that have these specific methylation patterns. These inhibitors are currently in clinical trials for solid tumors (12). Fukami et al. (5) suggests that therapy can be developed to target PPM1D, which is found to be overexpressed in both WHO grade II and grade III tumors, with the small molecule of SPI-001 that is currently used to treat breast cancers with mutations in PPM1D. However, this mutation was not measured in this study and has not yet been associated with any of the methylation classes at this time.
Finally, WHO grade I tumors were found in methylation classes of MC ben-1 (benign), MC ben-2 (benign), MC ben-3 (benign) and MC int-B (Intermediate). Patients with meningiomas in MC int-B were found to have a 23.1% probability of being progression-free at 72 months compared to patients with meningiomas in MC ben-1, MC ben-2 and MC ben-3 that were found to have a 75.7%, 83.9%, 88.9% probability of being progression-free at 72 months respectively. This approach at identifying the prognoses of WHO grade I tumors will be useful for clinicians in the process of considering patients for either observation or adjuvant treatment groups if resection can potentially be avoided. These methylation classes can help direct targeted therapy for WHO grade I tumors such as the use of AKT-inhibitor in the MC ben-2 methylation class as future targeted treatment options become available.
Though this was a fairly robust study with a large number of samples, it does have some limitations. These samples were retrospectively analyzed against their outcomes and a prospective study that utilized this new classification scheme to guide decision making would need to be completed to validate this model. Newly discovered gene mutations such as PPM1D and the POLR2A were not analyzed in the gene analysis, as this study began after their discovery. Future studies should associate methylation classes with more of the recently discovered genetic mutations as well as prognostic outcomes in patients. The authors mention that only 228 of the 497 patients’ clinical cases and prognoses were examined since the rest of the patients lacked sufficient clinical data, and they also mention their team was only able to sequence 303 of the 497 meningiomas for the portion of the study comparing previously established gene mutations with methylation classes.
Regardless, this study proposes an innovative and more precise system for diagnosing and classification of meningiomas, and this study is an excellent foundation for utilizing methylation patterns and other genetic classification to guide the treatment of meningiomas. DNA methylation profiles have already been defined for other CNS tumors such as ependymoma and glioma, and this has led to a greatly increased accuracy and efficiency of their diagnosis. Along with the recent literature of genetic mutations in meningiomas, these methylation classes help advance the process of diagnosing meningiomas from a strictly histological approach to one that incorporates both histology and molecular markers to deliver a more accurate treatment for meningiomas as recommended by the new 2016 WHO revision.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 2017;18:682-94. [Crossref] [PubMed]
- Bujko M, Machnicki MM, Grecka E, et al. Mutational Analysis of Recurrent Meningioma Progressing From Atypical to Rhabdoid Subtype. World Neurosurg 2017;97:754.e1-6. [Crossref] [PubMed]
- Yuzawa S, Nishihara H, Yamaguchi S, et al. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod Pathol 2016;29:708-16. [Crossref] [PubMed]
- van den Munckhof P, Christiaans I, Kenter SB, et al. Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the falx cerebri. Neurogenetics 2012;13:1-7. [Crossref] [PubMed]
- Fukami S, Riemenschneider MJ, Kohno M, et al. Expression and gene doses changes of the p53-regulator PPM1D in meningiomas: a role in meningioma progression? Brain Tumor Pathol 2016;33:191-9. [Crossref] [PubMed]
- Goutagny S, Nault JC, Mallet M, et al. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol 2014;24:184-9. [Crossref] [PubMed]
- Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet 2016;48:1253-9. [Crossref] [PubMed]
- Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol 2016;18:649-55. [Crossref] [PubMed]
- Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol 2013;125:351-8. [Crossref] [PubMed]
- Goldbrunner R, Minniti G, Preusser M, et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol 2016;17:e383-91. [Crossref] [PubMed]
- Shah NR, Tancioni I, Ward KK, et al. Analyses of merlin/NF2 connection to FAK inhibitor responsiveness in serous ovarian cancer. Gynecol Oncol 2014;134:104-11. [Crossref] [PubMed]
- Rimkus TK, Carpenter RL, Qasem S, et al. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 2016;8:E22. [Crossref] [PubMed]
Cite this article as: Gendreau JL, Chow KK, Sussman ES, Iyer A, Pendharkar AV, Ho AL. DNA methylation analysis for the treatment of meningiomas. J Vis Surg 2017;3:178.


